Note
Go to the end to download the full example code
Benchmark SamplesLoss in 3D
Let’s compare the performances of our losses and backends as the number of samples grows from 100 to 1,000,000.
Setup
import numpy as np
import time
from matplotlib import pyplot as plt
import importlib
import torch
use_cuda = torch.cuda.is_available()
from geomloss import SamplesLoss
MAXTIME = 10 if use_cuda else 1 # Max number of seconds before we break the loop
REDTIME = (
2 if use_cuda else 0.2
) # Decrease the number of runs if computations take longer than 2s...
D = 3 # Let's do this in 3D
# Number of samples that we'll loop upon
NS = [
100,
200,
500,
1000,
2000,
5000,
10000,
20000,
50000,
100000,
200000,
500000,
1000000,
]
Synthetic dataset. Feel free to use a Stanford Bunny, or whatever!
def generate_samples(N, device):
"""Create point clouds sampled non-uniformly on a sphere of diameter 1."""
x = torch.randn(N, D, device=device)
x[:, 0] += 1
x = x / (2 * x.norm(dim=1, keepdim=True))
y = torch.randn(N, D, device=device)
y[:, 1] += 2
y = y / (2 * y.norm(dim=1, keepdim=True))
x.requires_grad = True
# Draw random weights:
a = torch.randn(N, device=device)
b = torch.randn(N, device=device)
# And normalize them:
a = a.abs()
b = b.abs()
a = a / a.sum()
b = b / b.sum()
return a, x, b, y
Benchmarking loops.
def benchmark(Loss, dev, N, loops=10):
"""Times a loss computation+gradient on an N-by-N problem."""
# NB: torch does not accept reloading anymore.
# importlib.reload(torch) # In case we had a memory overflow just before...
device = torch.device(dev)
a, x, b, y = generate_samples(N, device)
# We simply benchmark a Loss + gradien wrt. x
code = "L = Loss( a, x, b, y ) ; L.backward()"
Loss.verbose = True
exec(code, locals()) # Warmup run, to compile and load everything
Loss.verbose = False
t_0 = time.perf_counter() # Actual benchmark --------------------
if use_cuda:
torch.cuda.synchronize()
for i in range(loops):
exec(code, locals())
if use_cuda:
torch.cuda.synchronize()
elapsed = time.perf_counter() - t_0 # ---------------------------
print(
"{:3} NxN loss, with N ={:7}: {:3}x{:3.6f}s".format(
loops, N, loops, elapsed / loops
)
)
return elapsed / loops
def bench_config(Loss, dev):
"""Times a loss computation+gradient for an increasing number of samples."""
print("Backend : {}, Device : {} -------------".format(Loss.backend, dev))
times = []
def run_bench():
try:
Nloops = [100, 10, 1]
nloops = Nloops.pop(0)
for n in NS:
elapsed = benchmark(Loss, dev, n, loops=nloops)
times.append(elapsed)
if (nloops * elapsed > MAXTIME) or (
nloops * elapsed > REDTIME and len(Nloops) > 0
):
nloops = Nloops.pop(0)
except IndexError:
print("**\nToo slow !")
try:
run_bench()
except RuntimeError as err:
if str(err)[:4] == "CUDA":
print("**\nMemory overflow !")
else:
# CUDA memory overflows semi-break the internal
# torch state and may cause some strange bugs.
# In this case, best option is simply to re-launch
# the benchmark.
run_bench()
return times + (len(NS) - len(times)) * [np.nan]
def full_bench(loss, *args, **kwargs):
"""Benchmarks the varied backends of a geometric loss function."""
print("Benchmarking : ===============================")
lines = [NS]
backends = ["tensorized", "online", "multiscale"]
for backend in backends:
Loss = SamplesLoss(*args, **kwargs, backend=backend)
lines.append(bench_config(Loss, "cuda" if use_cuda else "cpu"))
benches = np.array(lines).T
# Creates a pyplot figure:
plt.figure()
linestyles = ["o-", "s-", "^-"]
for i, backend in enumerate(backends):
plt.plot(
benches[:, 0],
benches[:, i + 1],
linestyles[i],
linewidth=2,
label='backend="{}"'.format(backend),
)
plt.title('Runtime for SamplesLoss("{}") in dimension {}'.format(Loss.loss, D))
plt.xlabel("Number of samples per measure")
plt.ylabel("Seconds")
plt.yscale("log")
plt.xscale("log")
plt.legend(loc="upper left")
plt.grid(True, which="major", linestyle="-")
plt.grid(True, which="minor", linestyle="dotted")
plt.axis([NS[0], NS[-1], 1e-3, MAXTIME])
plt.tight_layout()
# Save as a .csv to put a nice Tikz figure in the papers:
header = "Npoints " + " ".join(backends)
np.savetxt(
"output/benchmark_" + Loss.loss + "_3D.csv",
benches,
fmt="%-9.5f",
header=header,
comments="",
)
Gaussian MMD, with a small blur
full_bench(SamplesLoss, "gaussian", blur=0.1, truncate=3)
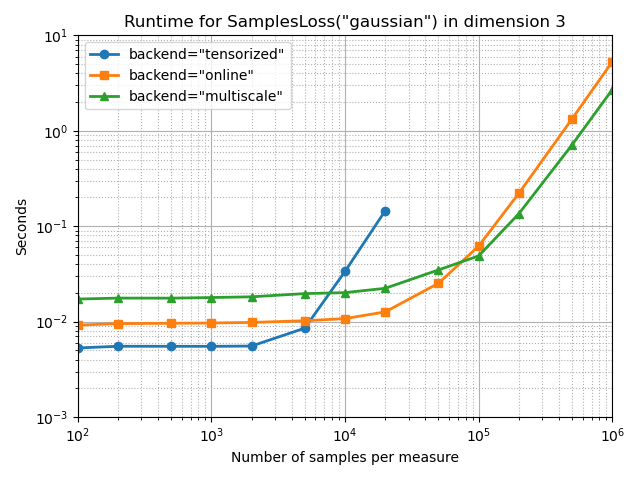
Benchmarking : ===============================
Backend : tensorized, Device : cuda -------------
100 NxN loss, with N = 100: 100x0.005299s
100 NxN loss, with N = 200: 100x0.005526s
100 NxN loss, with N = 500: 100x0.005521s
100 NxN loss, with N = 1000: 100x0.005516s
100 NxN loss, with N = 2000: 100x0.005554s
100 NxN loss, with N = 5000: 100x0.008550s
100 NxN loss, with N = 10000: 100x0.033574s
10 NxN loss, with N = 20000: 10x0.145354s
**
Memory overflow !
Backend : online, Device : cuda -------------
100 NxN loss, with N = 100: 100x0.009200s
100 NxN loss, with N = 200: 100x0.009534s
100 NxN loss, with N = 500: 100x0.009619s
100 NxN loss, with N = 1000: 100x0.009683s
100 NxN loss, with N = 2000: 100x0.009820s
100 NxN loss, with N = 5000: 100x0.010222s
100 NxN loss, with N = 10000: 100x0.010767s
100 NxN loss, with N = 20000: 100x0.012666s
100 NxN loss, with N = 50000: 100x0.025210s
10 NxN loss, with N = 100000: 10x0.061954s
10 NxN loss, with N = 200000: 10x0.221864s
1 NxN loss, with N = 500000: 1x1.335791s
1 NxN loss, with N =1000000: 1x5.302837s
Backend : multiscale, Device : cuda -------------
82x69 clusters, computed at scale = 0.782
100 NxN loss, with N = 100: 100x0.017255s
145x113 clusters, computed at scale = 0.781
100 NxN loss, with N = 200: 100x0.017660s
266x173 clusters, computed at scale = 0.786
100 NxN loss, with N = 500: 100x0.017636s
378x224 clusters, computed at scale = 0.791
100 NxN loss, with N = 1000: 100x0.017902s
486x306 clusters, computed at scale = 0.792
100 NxN loss, with N = 2000: 100x0.018219s
591x408 clusters, computed at scale = 0.793
100 NxN loss, with N = 5000: 100x0.019684s
640x465 clusters, computed at scale = 0.793
100 NxN loss, with N = 10000: 100x0.020147s
657x535 clusters, computed at scale = 0.794
10 NxN loss, with N = 20000: 10x0.022367s
692x618 clusters, computed at scale = 0.794
10 NxN loss, with N = 50000: 10x0.034833s
702x651 clusters, computed at scale = 0.794
10 NxN loss, with N = 100000: 10x0.048888s
712x674 clusters, computed at scale = 0.794
10 NxN loss, with N = 200000: 10x0.135483s
720x694 clusters, computed at scale = 0.794
10 NxN loss, with N = 500000: 10x0.713417s
725x707 clusters, computed at scale = 0.794
1 NxN loss, with N =1000000: 1x2.705595s
Energy Distance MMD
full_bench(SamplesLoss, "energy")
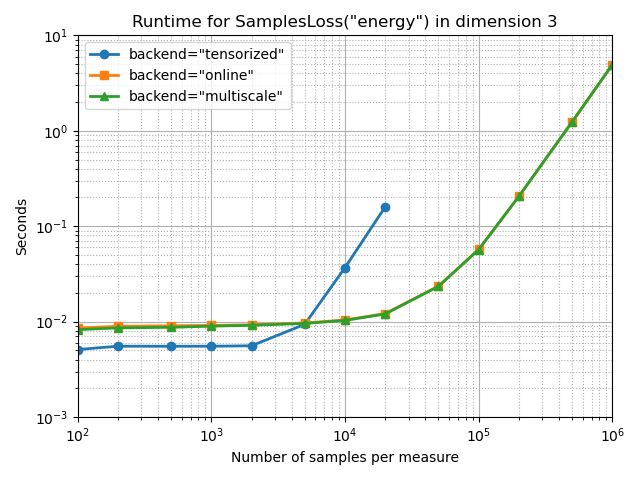
Benchmarking : ===============================
Backend : tensorized, Device : cuda -------------
100 NxN loss, with N = 100: 100x0.005103s
100 NxN loss, with N = 200: 100x0.005535s
100 NxN loss, with N = 500: 100x0.005526s
100 NxN loss, with N = 1000: 100x0.005533s
100 NxN loss, with N = 2000: 100x0.005587s
100 NxN loss, with N = 5000: 100x0.009383s
100 NxN loss, with N = 10000: 100x0.036845s
10 NxN loss, with N = 20000: 10x0.159529s
**
Memory overflow !
Backend : online, Device : cuda -------------
100 NxN loss, with N = 100: 100x0.008582s
100 NxN loss, with N = 200: 100x0.008939s
100 NxN loss, with N = 500: 100x0.009056s
100 NxN loss, with N = 1000: 100x0.009137s
100 NxN loss, with N = 2000: 100x0.009277s
100 NxN loss, with N = 5000: 100x0.009685s
100 NxN loss, with N = 10000: 100x0.010421s
100 NxN loss, with N = 20000: 100x0.012155s
100 NxN loss, with N = 50000: 100x0.023538s
10 NxN loss, with N = 100000: 10x0.057045s
10 NxN loss, with N = 200000: 10x0.205460s
1 NxN loss, with N = 500000: 1x1.239322s
1 NxN loss, with N =1000000: 1x4.925361s
Backend : multiscale, Device : cuda -------------
100 NxN loss, with N = 100: 100x0.008248s
100 NxN loss, with N = 200: 100x0.008605s
100 NxN loss, with N = 500: 100x0.008733s
100 NxN loss, with N = 1000: 100x0.008973s
100 NxN loss, with N = 2000: 100x0.009160s
100 NxN loss, with N = 5000: 100x0.009586s
100 NxN loss, with N = 10000: 100x0.010308s
100 NxN loss, with N = 20000: 100x0.012044s
100 NxN loss, with N = 50000: 100x0.023436s
10 NxN loss, with N = 100000: 10x0.056971s
10 NxN loss, with N = 200000: 10x0.205727s
1 NxN loss, with N = 500000: 1x1.238967s
1 NxN loss, with N =1000000: 1x4.933789s
Sinkhorn divergence
With a medium blurring scale, at one twentieth of the configuration’s diameter:
full_bench(SamplesLoss, "sinkhorn", p=2, blur=0.05, diameter=1)
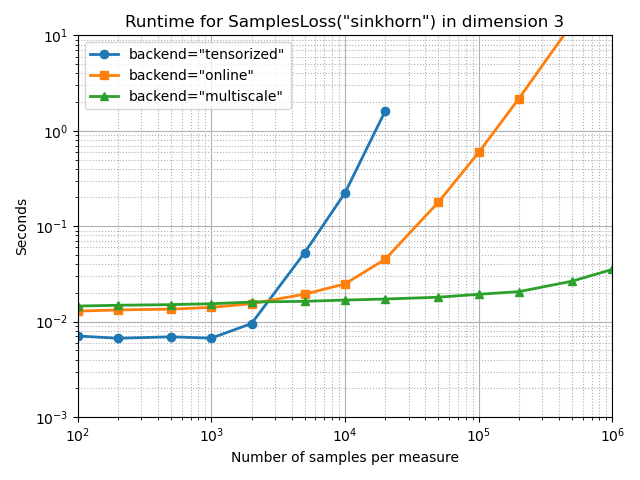
Benchmarking : ===============================
Backend : tensorized, Device : cuda -------------
100 NxN loss, with N = 100: 100x0.007079s
100 NxN loss, with N = 200: 100x0.006703s
100 NxN loss, with N = 500: 100x0.006941s
100 NxN loss, with N = 1000: 100x0.006722s
100 NxN loss, with N = 2000: 100x0.009562s
100 NxN loss, with N = 5000: 100x0.052947s
10 NxN loss, with N = 10000: 10x0.223527s
1 NxN loss, with N = 20000: 1x1.611537s
**
Memory overflow !
Backend : online, Device : cuda -------------
100 NxN loss, with N = 100: 100x0.012902s
100 NxN loss, with N = 200: 100x0.013262s
100 NxN loss, with N = 500: 100x0.013530s
100 NxN loss, with N = 1000: 100x0.014121s
100 NxN loss, with N = 2000: 100x0.015427s
100 NxN loss, with N = 5000: 100x0.019439s
100 NxN loss, with N = 10000: 100x0.024867s
10 NxN loss, with N = 20000: 10x0.045086s
10 NxN loss, with N = 50000: 10x0.178711s
10 NxN loss, with N = 100000: 10x0.593134s
1 NxN loss, with N = 200000: 1x2.175571s
1 NxN loss, with N = 500000: 1x13.285897s
**
Too slow !
Backend : multiscale, Device : cuda -------------
91x92 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 100: 100x0.014569s
181x159 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 200: 100x0.014862s
384x301 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 500: 100x0.015084s
633x424 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 1000: 100x0.015415s
919x595 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 2000: 100x0.016064s
1327x847 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 5000: 100x0.016333s
1632x1031 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 10000: 100x0.016842s
1832x1247 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 20000: 100x0.017265s
2003x1563 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 50000: 100x0.018045s
2073x1765 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 100000: 100x0.019364s
2124x1910 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
100 NxN loss, with N = 200000: 100x0.020637s
2172x2053 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
10 NxN loss, with N = 500000: 10x0.026567s
2185x2113 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.050
Extrapolate from coarse to fine after the last iteration.
10 NxN loss, with N =1000000: 10x0.035292s
With a small blurring scale, at one hundredth of the configuration’s diameter:
full_bench(SamplesLoss, "sinkhorn", p=2, blur=0.01, diameter=1)
plt.show()
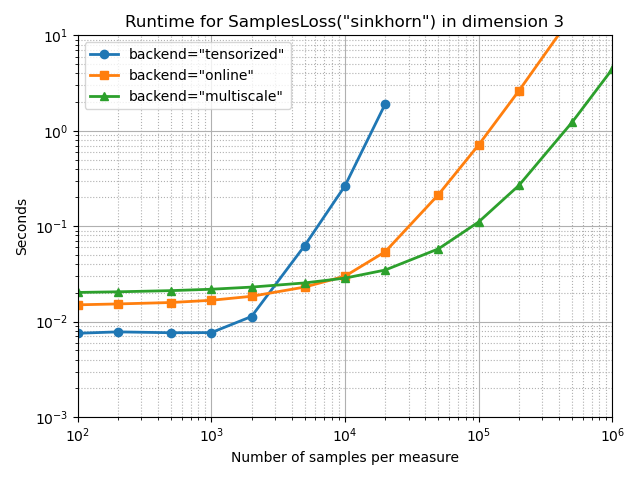
Benchmarking : ===============================
Backend : tensorized, Device : cuda -------------
100 NxN loss, with N = 100: 100x0.007561s
100 NxN loss, with N = 200: 100x0.007817s
100 NxN loss, with N = 500: 100x0.007656s
100 NxN loss, with N = 1000: 100x0.007682s
100 NxN loss, with N = 2000: 100x0.011329s
100 NxN loss, with N = 5000: 100x0.062733s
10 NxN loss, with N = 10000: 10x0.264870s
1 NxN loss, with N = 20000: 1x1.909170s
**
Memory overflow !
Backend : online, Device : cuda -------------
100 NxN loss, with N = 100: 100x0.015015s
100 NxN loss, with N = 200: 100x0.015341s
100 NxN loss, with N = 500: 100x0.015877s
100 NxN loss, with N = 1000: 100x0.016754s
100 NxN loss, with N = 2000: 100x0.018445s
100 NxN loss, with N = 5000: 100x0.023053s
10 NxN loss, with N = 10000: 10x0.029969s
10 NxN loss, with N = 20000: 10x0.054050s
10 NxN loss, with N = 50000: 10x0.214230s
1 NxN loss, with N = 100000: 1x0.711520s
1 NxN loss, with N = 200000: 1x2.624156s
1 NxN loss, with N = 500000: 1x16.047510s
**
Too slow !
Backend : multiscale, Device : cuda -------------
96x81 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 1273/7776 = 16.4% of the coarse cost matrix.
Keep 1208/9216 = 13.1% of the coarse cost matrix.
Keep 1793/6561 = 27.3% of the coarse cost matrix.
100 NxN loss, with N = 100: 100x0.020246s
178x159 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 3885/28302 = 13.7% of the coarse cost matrix.
Keep 3754/31684 = 11.8% of the coarse cost matrix.
Keep 5067/25281 = 20.0% of the coarse cost matrix.
10 NxN loss, with N = 200: 10x0.020553s
379x295 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 14892/111805 = 13.3% of the coarse cost matrix.
Keep 15537/143641 = 10.8% of the coarse cost matrix.
Keep 16697/87025 = 19.2% of the coarse cost matrix.
10 NxN loss, with N = 500: 10x0.021149s
630x435 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 34578/274050 = 12.6% of the coarse cost matrix.
Keep 40168/396900 = 10.1% of the coarse cost matrix.
Keep 32093/189225 = 17.0% of the coarse cost matrix.
10 NxN loss, with N = 1000: 10x0.021868s
934x603 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 63295/563202 = 11.2% of the coarse cost matrix.
Keep 79926/872356 = 9.2% of the coarse cost matrix.
Keep 53179/363609 = 14.6% of the coarse cost matrix.
10 NxN loss, with N = 2000: 10x0.023026s
1359x843 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 115167/1145637 = 10.1% of the coarse cost matrix.
Keep 153739/1846881 = 8.3% of the coarse cost matrix.
Keep 85789/710649 = 12.1% of the coarse cost matrix.
10 NxN loss, with N = 5000: 10x0.025470s
1609x1044 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 162316/1679796 = 9.7% of the coarse cost matrix.
Keep 210177/2588881 = 8.1% of the coarse cost matrix.
Keep 118852/1089936 = 10.9% of the coarse cost matrix.
10 NxN loss, with N = 10000: 10x0.028603s
1858x1255 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 216719/2331790 = 9.3% of the coarse cost matrix.
Keep 275990/3452164 = 8.0% of the coarse cost matrix.
Keep 157385/1575025 = 10.0% of the coarse cost matrix.
10 NxN loss, with N = 20000: 10x0.034766s
2004x1562 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 279293/3130248 = 8.9% of the coarse cost matrix.
Keep 321418/4016016 = 8.0% of the coarse cost matrix.
Keep 224342/2439844 = 9.2% of the coarse cost matrix.
10 NxN loss, with N = 50000: 10x0.057913s
2065x1742 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 310590/3597230 = 8.6% of the coarse cost matrix.
Keep 341841/4264225 = 8.0% of the coarse cost matrix.
Keep 269452/3034564 = 8.9% of the coarse cost matrix.
10 NxN loss, with N = 100000: 10x0.110885s
2128x1927 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 345694/4100656 = 8.4% of the coarse cost matrix.
Keep 363590/4528384 = 8.0% of the coarse cost matrix.
Keep 326729/3713329 = 8.8% of the coarse cost matrix.
10 NxN loss, with N = 200000: 10x0.267706s
2166x2060 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 370323/4461960 = 8.3% of the coarse cost matrix.
Keep 377652/4691556 = 8.0% of the coarse cost matrix.
Keep 375630/4243600 = 8.9% of the coarse cost matrix.
1 NxN loss, with N = 500000: 1x1.228583s
2183x2115 clusters, computed at scale = 0.046
Successive scales : 1.000, 1.000, 0.500, 0.250, 0.125, 0.063, 0.031, 0.016, 0.010
Jump from coarse to fine between indices 5 (σ=0.063) and 6 (σ=0.031).
Keep 380326/4617045 = 8.2% of the coarse cost matrix.
Keep 383841/4765489 = 8.1% of the coarse cost matrix.
Keep 397855/4473225 = 8.9% of the coarse cost matrix.
1 NxN loss, with N =1000000: 1x4.436718s
Total running time of the script: (4 minutes 56.952 seconds)