Note
Go to the end to download the full example code
Kernel interpolation - NumPy API
The pykeops.numpy.LazyTensor.solve(b, alpha=1e-10) method of KeOps pykeops.numpy.LazyTensor allows you to solve optimization
problems of the form
where \(K_{xx}\) is a symmetric, positive definite linear operator defined through the KeOps generic syntax and \(\alpha\) is a nonnegative regularization parameter. In the following script, we use it to solve large-scale Kriging (aka. Gaussian process regression or generalized spline interpolation) problems with a linear memory footprint.
Setup
Standard imports:
import time
import numpy as np
from matplotlib import pyplot as plt
from pykeops.numpy import LazyTensor
import pykeops.config
Generate some data:
dtype = "float64"
N = 10000 if pykeops.config.gpu_available else 1000 # Number of samples
# Sampling locations:
x = np.random.rand(N, 1).astype(dtype)
# Some random-ish 1D signal:
b = (
x
+ 0.5 * np.sin(6 * x)
+ 0.1 * np.sin(20 * x)
+ 0.05 * np.random.randn(N, 1).astype(dtype)
)
Interpolation in 1D
Specify our regression model - a simple Gaussian variogram or kernel matrix of deviation sigma:
def gaussian_kernel(x, y, sigma=0.1):
x_i = LazyTensor(x[:, None, :]) # (M, 1, 1)
y_j = LazyTensor(y[None, :, :]) # (1, N, 1)
D_ij = ((x_i - y_j) ** 2).sum(-1) # (M, N) symbolic matrix of squared distances
return (-D_ij / (2 * sigma**2)).exp() # (M, N) symbolic Gaussian kernel matrix
Perform the Kernel interpolation, without forgetting to specify the ridge regularization parameter alpha which controls the trade-off between a perfect fit (alpha = 0) and a smooth interpolation (alpha = \(+\infty\)):
alpha = 1.0 # Ridge regularization
start = time.time()
K_xx = gaussian_kernel(x, x)
a = K_xx.solve(b, alpha=alpha)
end = time.time()
print(
"Time to perform an RBF interpolation with {:,} samples in 1D: {:.5f}s".format(
N, end - start
)
)
Time to perform an RBF interpolation with 10,000 samples in 1D: 0.45912s
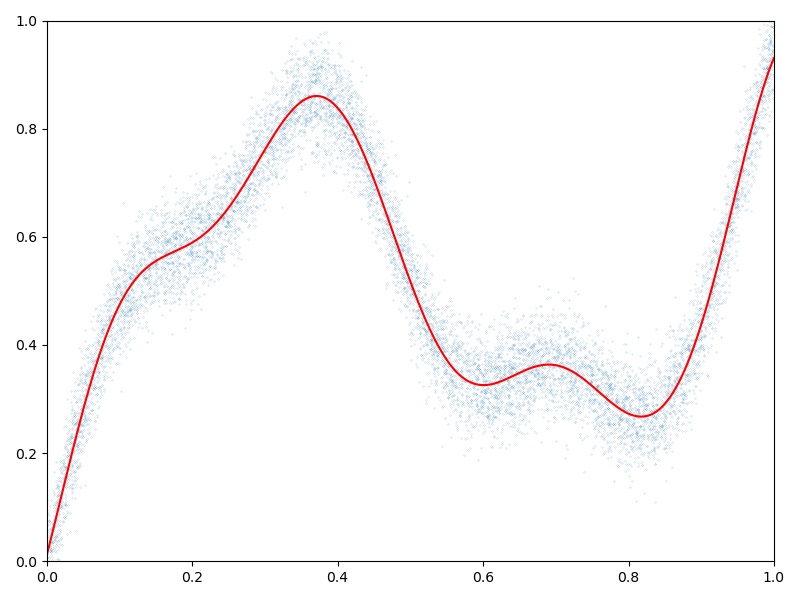
Display the (fitted) model on the unit interval:
# Extrapolate on a uniform sample:
t = np.reshape(np.linspace(0, 1, 1001), [1001, 1]).astype(dtype)
K_tx = gaussian_kernel(t, x)
mean_t = K_tx @ a
# 1D plot:
plt.figure(figsize=(8, 6))
plt.scatter(x[:, 0], b[:, 0], s=100 / len(x)) # Noisy samples
plt.plot(t, mean_t, "r")
plt.axis([0, 1, 0, 1])
plt.tight_layout()
Interpolation in 2D
Generate some data:
# Sampling locations:
x = np.random.rand(N, 2).astype(dtype)
# Some random-ish 2D signal:
b = np.sum((x - 0.5) ** 2, axis=1)[:, None]
b[b > 0.4**2] = 0
b[b < 0.3**2] = 0
b[b >= 0.3**2] = 1
b = b + 0.05 * np.random.randn(N, 1).astype(dtype)
# Add 25% of outliers:
Nout = N // 4
b[-Nout:] = np.random.rand(Nout, 1).astype(dtype)
Specify our regression model - a simple Exponential variogram or Laplacian kernel matrix of deviation sigma:
def laplacian_kernel(x, y, sigma=0.1):
x_i = LazyTensor(x[:, None, :]) # (M, 1, 1)
y_j = LazyTensor(y[None, :, :]) # (1, N, 1)
D_ij = ((x_i - y_j) ** 2).sum(-1) # (M, N) symbolic matrix of squared distances
return (-D_ij.sqrt() / sigma).exp() # (M, N) symbolic Laplacian kernel matrix
Perform the Kernel interpolation, without forgetting to specify the ridge regularization parameter alpha which controls the trade-off between a perfect fit (alpha = 0) and a smooth interpolation (alpha = \(+\infty\)):
alpha = 10 # Ridge regularization
start = time.time()
K_xx = laplacian_kernel(x, x)
a = K_xx.solve(b, alpha=alpha)
end = time.time()
print(
"Time to perform an RBF interpolation with {:,} samples in 2D: {:.5f}s".format(
N, end - start
)
)
Time to perform an RBF interpolation with 10,000 samples in 2D: 0.74956s
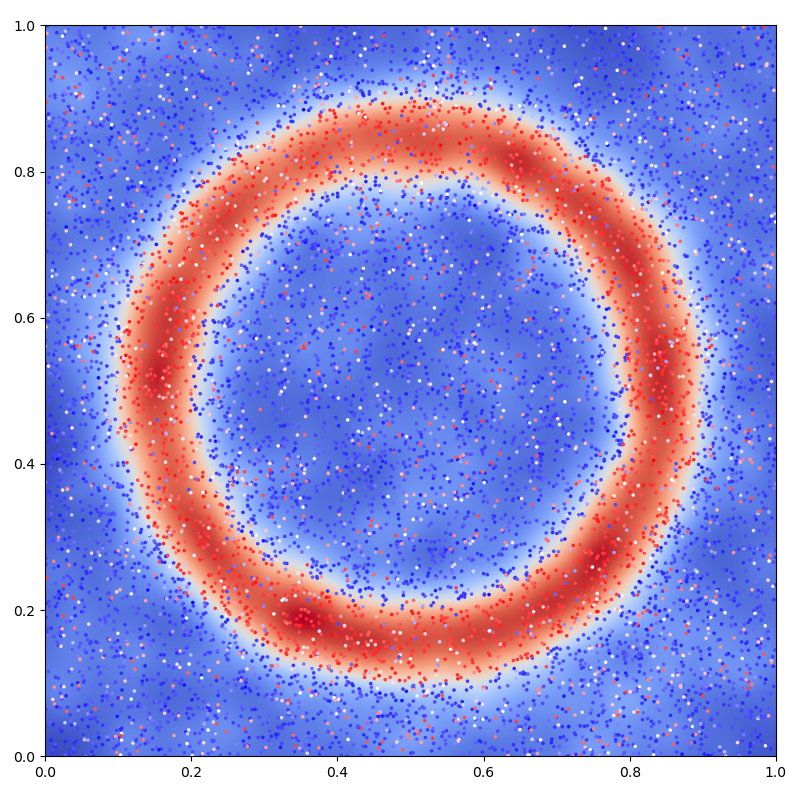
Display the (fitted) model on the unit square:
# Extrapolate on a uniform sample:
X = Y = np.linspace(0, 1, 101)
X, Y = np.meshgrid(X, Y)
t = np.stack((X.ravel(), Y.ravel()), axis=1)
K_tx = laplacian_kernel(t, x)
mean_t = K_tx @ a
mean_t = mean_t.reshape(101, 101)[::-1, :]
# 2D plot: noisy samples and interpolation in the background
plt.figure(figsize=(8, 8))
plt.scatter(x[:, 0], x[:, 1], c=b.ravel(), s=25000 / len(x), cmap="bwr")
plt.imshow(mean_t, interpolation="bilinear", extent=[0, 1, 0, 1], cmap="coolwarm")
plt.axis([0, 1, 0, 1])
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 1.621 seconds)