Note
Click here to download the full example code
Unstructured Deformations to Model Acropetal Growth¶
Import relevant Python modules.
import sys
sys.path.append("../../")
import math
import copy
import pickle
import torch
print(torch.__version__)
import matplotlib.pyplot as plt
import imodal
torch.set_default_dtype(torch.float32)
imodal.Utilities.set_compute_backend('torch')
Out:
1.3.0
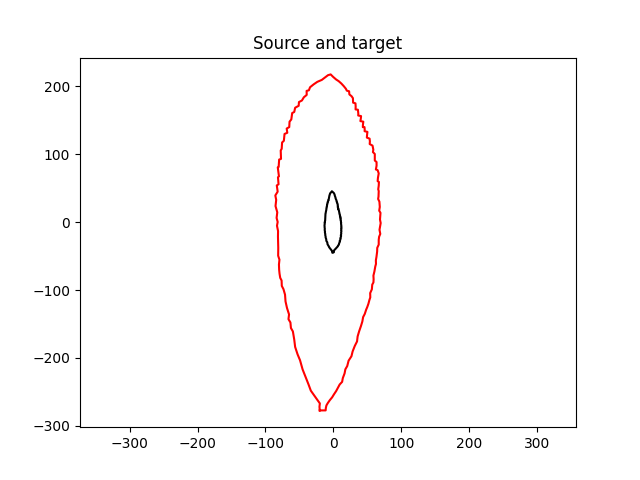
We load the data (shape of the source and target leaves).
with open("../../data/acropetal.pickle", 'rb') as f:
data = pickle.load(f)
shape_source = imodal.Utilities.close_shape(torch.tensor(data['shape_source']).type(torch.get_default_dtype()))
shape_target = imodal.Utilities.close_shape(torch.tensor(data['shape_target']).type(torch.get_default_dtype()))
aabb_source = imodal.Utilities.AABB.build_from_points(shape_source)
aabb_target = imodal.Utilities.AABB.build_from_points(shape_target)
Plot source and target.
plt.title("Source and target")
plt.plot(shape_source[:, 0].numpy(), shape_source[:, 1].numpy(), color='black')
plt.plot(shape_target[:, 0].numpy(), shape_target[:, 1].numpy(), color='red')
plt.axis('equal')
plt.show()
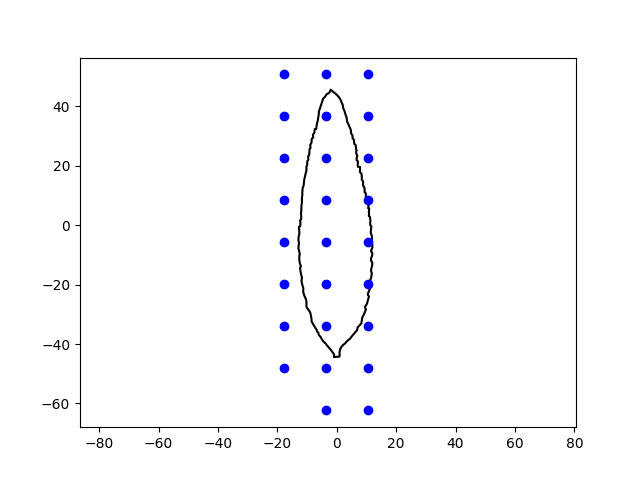
We now sample the points that will be used by the implicit deformation module of order 0 (LDDMM module).
# Build AABB (Axis Aligned Bounding Box) around the source shape and uniformly
# sample points for the growth module.
points_density = 0.005
points_translations = imodal.Utilities.fill_area_uniform_density(imodal.Utilities.area_shape, aabb_source.scale(1.4), points_density, shape=2.*shape_source)
Plot points of the local translations module.
plt.plot(shape_source[:, 0].numpy(), shape_source[:, 1].numpy(), color='black')
plt.plot(points_translations[:, 0].numpy(), points_translations[:, 1].numpy(), 'o', color='blue')
plt.axis('equal')
plt.show()
Create the deformation model which only consists of one implicit module of order 0.
Create and initialize implicit module of order 0.
sigma = 4./points_density**(1/2)
print(sigma)
translations = imodal.DeformationModules.Translations(2, points_translations.shape[0], sigma, gd=points_translations)
Out:
56.568542494923804
Define deformables used by the registration model.
deformable_shape_source = imodal.Models.DeformablePoints(shape_source)
deformable_shape_target = imodal.Models.DeformablePoints(shape_target)
Registration¶
Define the registration model.
model = imodal.Models.RegistrationModel(
[deformable_shape_source],
[translations],
[imodal.Attachment.VarifoldAttachment(2, [50., 300.])],
lam=10.)
Fitting using Torch LBFGS optimizer.
shoot_solver = 'euler'
shoot_it = 10
costs = {}
fitter = imodal.Models.Fitter(model, optimizer='torch_lbfgs')
fitter.fit([deformable_shape_target], 50, costs=costs, options={'shoot_solver': shoot_solver, 'shoot_it': shoot_it, 'line_search_fn': 'strong_wolfe'})
Out:
Starting optimization with method torch LBFGS, using solver euler with 10 iterations.
Initial cost={'deformation': 0.0, 'attach': 5747971.0}
1e-10
Evaluated model with costs=5747971.0
Evaluated model with costs=5735429.089713693
Evaluated model with costs=5678879.727811813
Evaluated model with costs=5061769.626953125
Evaluated model with costs=3262514.7250976562
Evaluated model with costs=3506628.03125
Evaluated model with costs=1950700.359375
Evaluated model with costs=1639311.9453125
Evaluated model with costs=53068309331968.0
Evaluated model with costs=1621987.48046875
Evaluated model with costs=1621954.484375
Evaluated model with costs=1607418.220703125
Evaluated model with costs=1449339.72265625
Evaluated model with costs=1371464.99609375
Evaluated model with costs=1292304.08984375
Evaluated model with costs=1292437.08203125
Evaluated model with costs=1263735.3671875
Evaluated model with costs=1231730.65234375
Evaluated model with costs=1210437.81640625
Evaluated model with costs=1170838.796875
Evaluated model with costs=1110834.19921875
Evaluated model with costs=991232.34375
Evaluated model with costs=958222.07421875
Evaluated model with costs=791807.0703125
Evaluated model with costs=5145467.341796875
Evaluated model with costs=805486.3984375
Evaluated model with costs=622646.2734375
================================================================================
Time: 7.4879589710001255
Iteration: 0
Costs
deformation=51364.5234375
attach=571281.75
Total cost=622646.2734375
1e-10
Evaluated model with costs=622646.2734375
Evaluated model with costs=523710.203125
Evaluated model with costs=364900.26953125
Evaluated model with costs=308105.8125
Evaluated model with costs=257276.5
Evaluated model with costs=243784.0390625
Evaluated model with costs=190984.953125
Evaluated model with costs=155093.24609375
Evaluated model with costs=150918.47265625
Evaluated model with costs=167951.515625
Evaluated model with costs=150127.3671875
Evaluated model with costs=136170.37890625
Evaluated model with costs=112797.1015625
Evaluated model with costs=106412.5
Evaluated model with costs=95385.359375
Evaluated model with costs=90303.203125
Evaluated model with costs=89506.69921875
Evaluated model with costs=88359.21484375
Evaluated model with costs=87437.7890625
Evaluated model with costs=550377.546875
Evaluated model with costs=86746.5
Evaluated model with costs=22689564.875
Evaluated model with costs=87265.8671875
Evaluated model with costs=85442.453125
Evaluated model with costs=87675.2421875
Evaluated model with costs=95814.328125
Evaluated model with costs=87705.61328125
Evaluated model with costs=89252.05078125
Evaluated model with costs=85442.453125
Evaluated model with costs=85442.453125
================================================================================
Time: 15.810912368000118
Iteration: 1
Costs
deformation=52032.296875
attach=33410.15625
Total cost=85442.453125
1e-10
Evaluated model with costs=85442.453125
Evaluated model with costs=87675.2421875
Evaluated model with costs=95814.328125
Evaluated model with costs=87705.61328125
Evaluated model with costs=89252.05078125
Evaluated model with costs=85442.453125
Evaluated model with costs=85442.453125
================================================================================
Time: 17.72755951099998
Iteration: 2
Costs
deformation=52032.296875
attach=33410.15625
Total cost=85442.453125
================================================================================
Optimisation process exited with message: Convergence achieved.
Final cost=85442.453125
Model evaluation count=64
Time elapsed = 17.728239290000147
Visualization¶
Compute optimized deformation trajectory.
intermediates = {}
with torch.autograd.no_grad():
deformed = model.compute_deformed(shoot_solver, shoot_it, intermediates=intermediates)
deformed_shape = deformed[0][0]
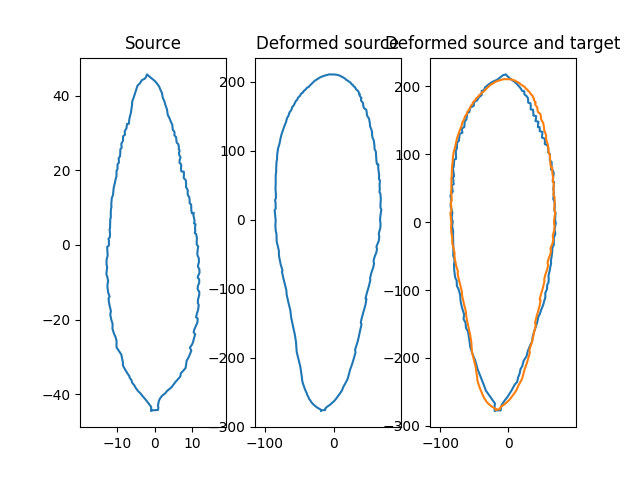
Plot results.
plt.subplot(1, 3, 1)
plt.title("Source")
plt.plot(shape_source[:, 0].numpy(), shape_source[:, 1].numpy(), '-')
plt.axis(aabb_target.totuple())
plt.axis('equal')
plt.subplot(1, 3, 2)
plt.title("Deformed source")
plt.plot(deformed_shape[:, 0], deformed_shape[:, 1], '-')
plt.axis(aabb_target.totuple())
plt.axis('equal')
plt.subplot(1, 3, 3)
plt.title("Deformed source and target")
plt.plot(shape_target[:, 0].numpy(), shape_target[:, 1].numpy(), '-')
plt.plot(deformed_shape[:, 0], deformed_shape[:, 1], '-')
plt.axis(aabb_target.totuple())
plt.axis('equal')
plt.show()
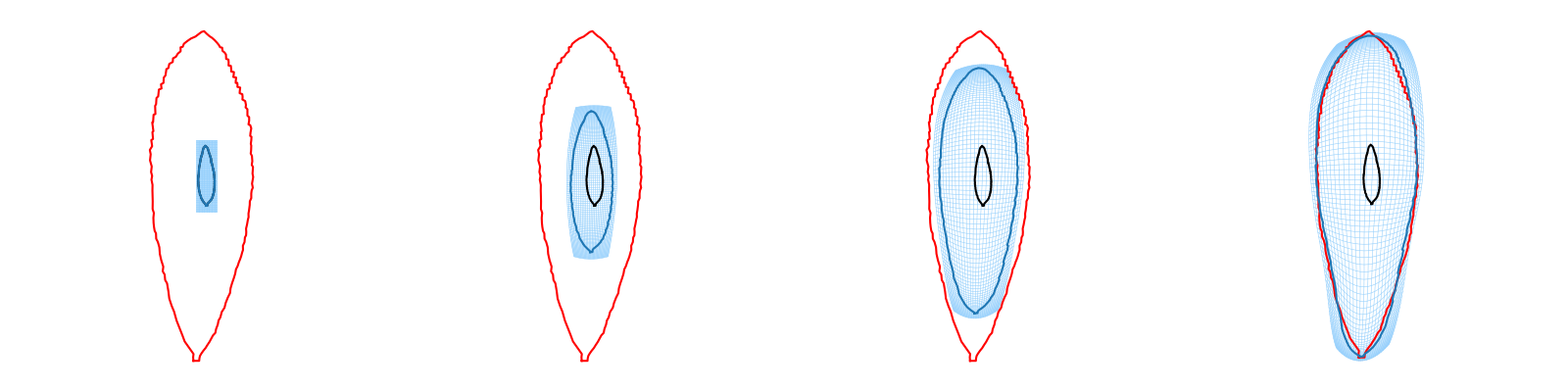
Recompute the learned deformation trajectory this time with the grid deformation to visualize growth.
# Reset the local translations module with the learned initialization manifold.
translations.manifold.fill(model.init_manifold[1])
aabb_source.scale_(1.2)
# Define the deformation grid.
square_size = 1
grid_resolution = [math.floor(aabb_source.width/square_size),
math.floor(aabb_source.height/square_size)]
deformable_source = imodal.Models.DeformablePoints(shape_source)
deformable_grid = imodal.Models.DeformableGrid(aabb_source, grid_resolution)
deformable_source.silent_module.manifold.fill_cotan(model.init_manifold[0].cotan)
controls = [[control[1]] for control in intermediates['controls']]
# Shoot.
intermediates = {}
with torch.autograd.no_grad():
imodal.Models.deformables_compute_deformed([deformable_source, deformable_grid], [translations], shoot_solver, shoot_it, controls=controls, intermediates=intermediates)
Plot the growth trajectory.
indices = [0, 3, 7, 10]
fig = plt.figure(figsize=[4.*len(indices), 4.])
for i, index in enumerate(indices):
state = intermediates['states'][index]
ax = plt.subplot(1, len(indices), i + 1)
deformable_grid.silent_module.manifold.fill_gd(state[1].gd)
grid_x, grid_y = deformable_grid.silent_module.togrid()
imodal.Utilities.plot_grid(ax, grid_x, grid_y, color='xkcd:light blue', lw=0.4)
plt.plot(shape_source[:, 0].numpy(), shape_source[:, 1].numpy(), color='black')
plt.plot(shape_target[:, 0].numpy(), shape_target[:, 1].numpy(), color='red')
plt.plot(state[0].gd[:, 0].numpy(), state[0].gd[:, 1].numpy())
plt.axis('equal')
plt.axis('off')
fig.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 19.596 seconds)